Making
CAS BioFinder
Challenge
Create a platform of solutions for medicinal chemists and biologists to search curated and connected data to make informed decisions regarding the drug-discovery process.
CAS BioFinder, a biological data discovery platform developed by CAS (a division of the American Chemical Society), is designed to assist scientists, particularly those in drug discovery, biomedical research, and life sciences, in discovering and connecting biological, chemical, and pharmacological information within a unified environment.
Impact
A 0 → 1, design and research driven product with over 1,500 users which delivers drug/target/disease relationships with 82% time-to-task completion for biologists and medicinal chemists, netting $3.67M additional revenue for the 2025 fiscal year.
My Role
Product designer, research lead, and eventually strategic lead (when assigned to a designer reporting to me).
Timeline
August 2022 - Present
Background & Research
CAS BioFinder started as a concept, codenamed “Darwin” in 2016 for an expansion into the biologics space for CAS content curation. A prototype was developed during that time and informed a lot of preliminary research which was inherited for this project.
CAS was also actively benchmarking existing software solutions to retrofit a framework and functionality to break into this space.
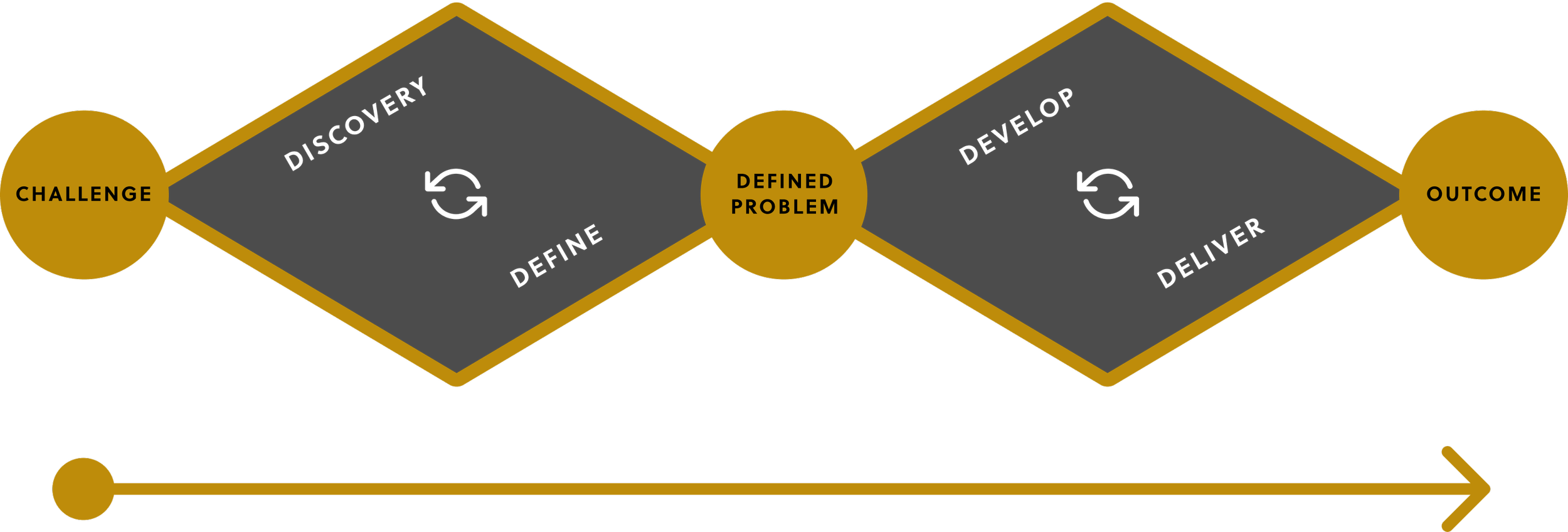
While vetting and discovering the features within a software called Chemotargets by Clarity, I began creating a framework for generative research. This software was developed by an academic firm and did not undergo any user research or design processes by the company. The UX program that I manage leverages the existing double diamond paradigm for discovery, definition, ideation, and implementation for all new and existing products.
Using the existing application, the Darwin concept from 2016, and similar product benchmarks, I designed low fidelity prototypes to convey organizational concepts and information for research candidates. The challenge was to find clean data which would be representative of the CAS content curation efforts. Scientists require factually accurate information to feel comfortable remarking on the efficacy of a prototype in research.
I collaborated on a viable search subject with colleagues from the content operations division and developed a clickable prototype with existing product design language elements in Adobe XD.
After commissioning and collaborating on research with an external agency, I participated in interview sessions with existing CAS customers and non-candidates who passed a rigorous screener survey regarding workflows and core application concepts. The research leveraged rapid iterative testing and evaluation (RITE) usability assessments to quickly address gaps in the design and workflow.
Competitive Analysis, Prototyping, & Conducting Research
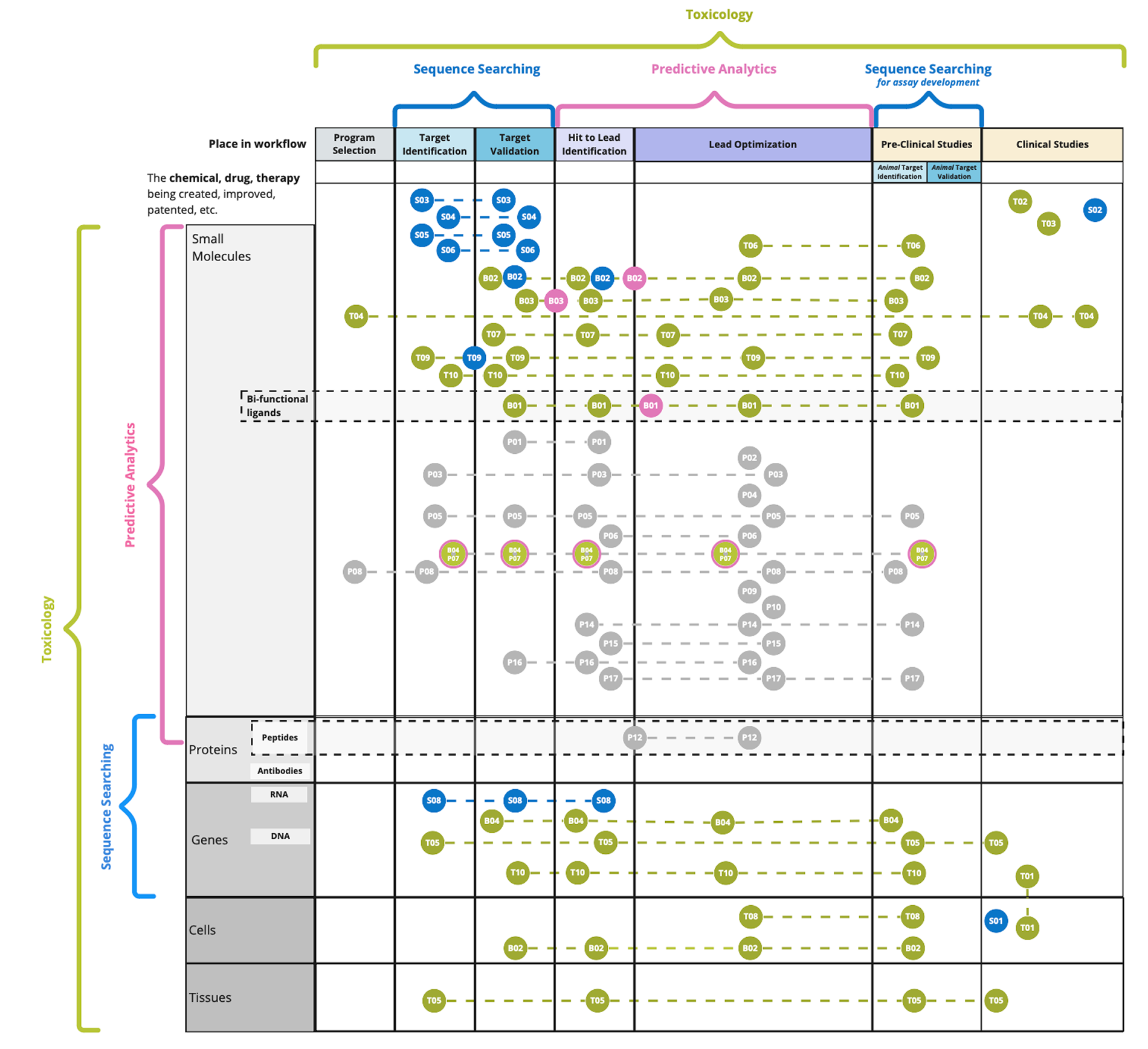
I created affinity maps which lead to the construction of the core personas for the new product: medicinal chemists and computational chemists. Other persona workflows were noted that will be implemented in the future which include toxicologists, molecular biologists, and biochemists.
Three key themes emerged from the analysis:
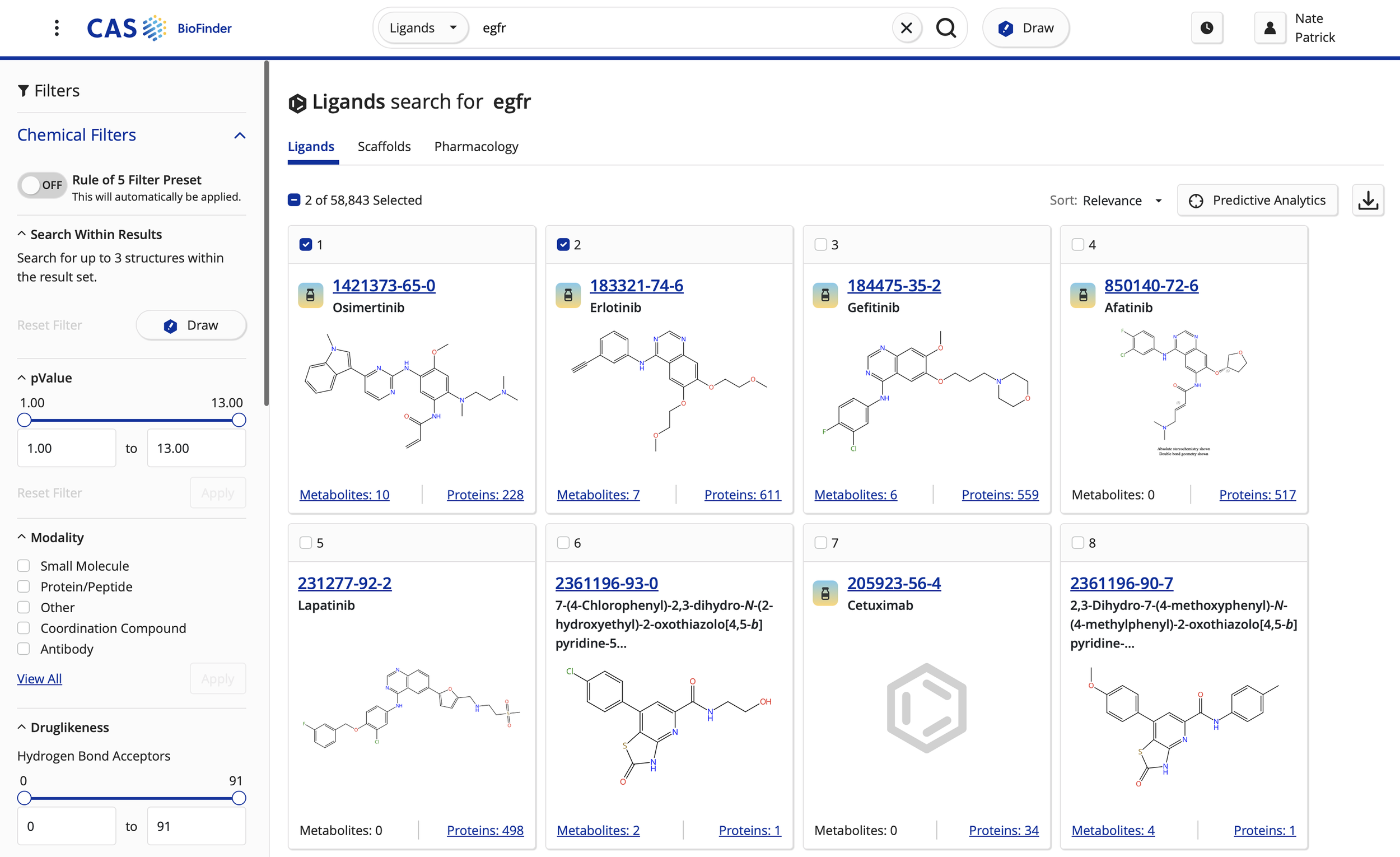
Customers wanted a one-stop-shop for all drug/ligand details and compare that information with binding affinity against known targets (proteins). Scaffold searching is a similar, albeit separate paradigm to group together similar drugs/ligands.
Referential information (journal and patents) was critical to the workflow to validate hunches and other information against searches for ligands/drugs, and their protein pairs.
The ability to upload proprietary molecules and algorithmically determine their predicted binding affinity was highly desired by customers which provided a unique challenge regarding the upload process and technical security of highly valuable intellectual property.
Process & Development
I worked directly with the product manager to prioritize epics of work which would create the core information architecture as defined and validated by the user research. In total, 215 epics were created for the commercial launch of the product. This visibility and level of planning provided guidance for engineering efforts and sizing of work.
I worked with the Director of Life Sciences and the Product Manager to craft a six quarter roadmap where the first four quarters were pre-commercial launch and then two more quarters of feature expansion to increase breadth and scale of the application post launch.
Working closely with the product manager, I created high-fidelity mockups and production-ready specifications for the UI in Figma. This was during the time our team transitioned from Adobe XD. I actively developed a product-specific design library and components to expedite the creation of specifications. The assets were delivered using Dev Mode in Figma, which the teams were delighted to learn how to use.
Transitioning from Adobe XD to Figma enabled more spontaneous transparency with the product and engineering teams. During this period, I closely collaborated with the front-end engineers to ensure tighter designs and user experience workflows, which I meticulously documented in JIRA cards. Before a new build was pushed to production in the Beta program, I partnered with QA to ensure that the designs were implemented to specification.
After 11 months of development, the application was launched commercially in May 2024. Throughout launch multiple meetings were conducted soliciting feedback to discover additional desired functionality based on beta program usage of the MVP version of BioFinder.
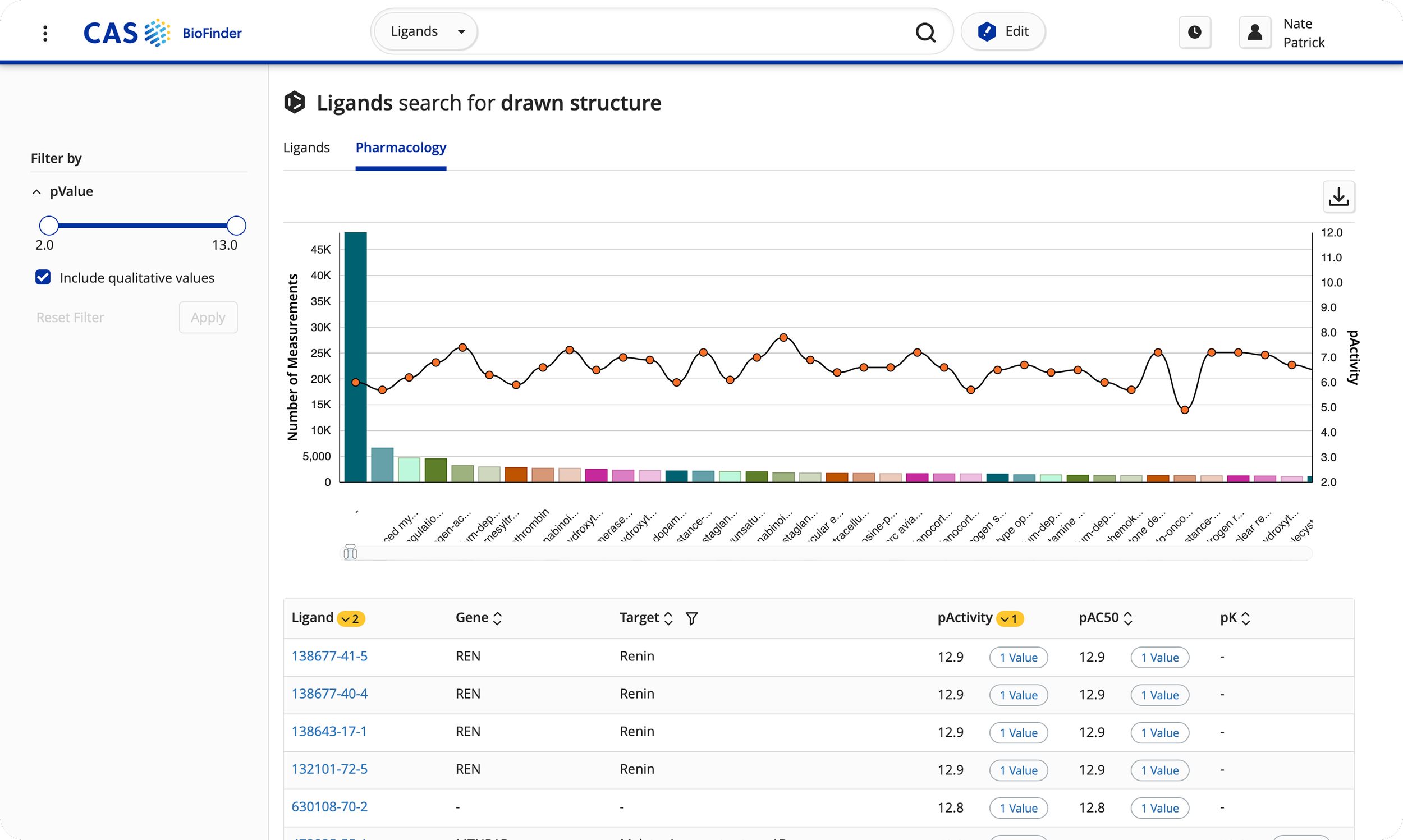
We also discovered that the vast majority of users (72.6%) preferred searching by drawn or known structures in lieu of text-based queries (27.4%).
Commercial Launch & Feedback
Customers were thrilled with the focused and tight solution we have delivered, and continue to want increased functionality to save time in the library and conduct/validate findings in the lab. Of the existing and potential customers surveyed, 82% said this solution is highly desirable to them and are excited to purchase when released in May of 2024.
“BioFinder is a very unique, interesting product which associates proteins and structures with assay data ... There are some other databases out there but I think this one is much more user friendly to chemists."
Biochemist Research Participant
It’s a really nice tool. … it can like summarize your literature search... So if you're working on a new project and you really want to get a lot of information in a short period of time, then … get an insight into the ligand or the specific target.”
Medicinal Chemist Research Participant
Learning & Adjustments
After launch, we monitored the usage of the product and kept tabs on sales motions daily. While the feedback was good, I began to craft a second version of drafts to adjust the UX to be more inline with what medicinal chemists and biochemists (our primary personas) would expect for integration of protein and disease detail pages.
Production adoption was slow as CAS’s primary sales mechanism was selling to chemists. Leadership determined that an open trial period would be drive growth and leverage our existing customers as advocates to their biological peers in organizations.—encouraging them to give CAS BioFinder a try in their daily research.
While our users are highly intelligent scientists, learning new tools can be overwhelming to them. It was during this time that the UX team vetted Appcues, an in-product messaging and onboarding tool, to create first-time-onboarding flows and explain new features and how customers could derive the most value out of their experience.
Refining & Releasing
Monitoring users activities and continually listening to feedback in research sessions moderated by me, I compiled a list of features and enhancements which would both feed the roadmap, and accelerate development of tools that would satisfy a third user persona: the biologist.
The first release of BioFinder was intended to hook the interest of familiar customers in chemists who worked in the pharmaceutical space. The second major release added the following functionality:
Development of two additional search types: proteins and diseases. Tying all search types together to allow users to circumnavigate the application from anywhere in their research or development workflow.
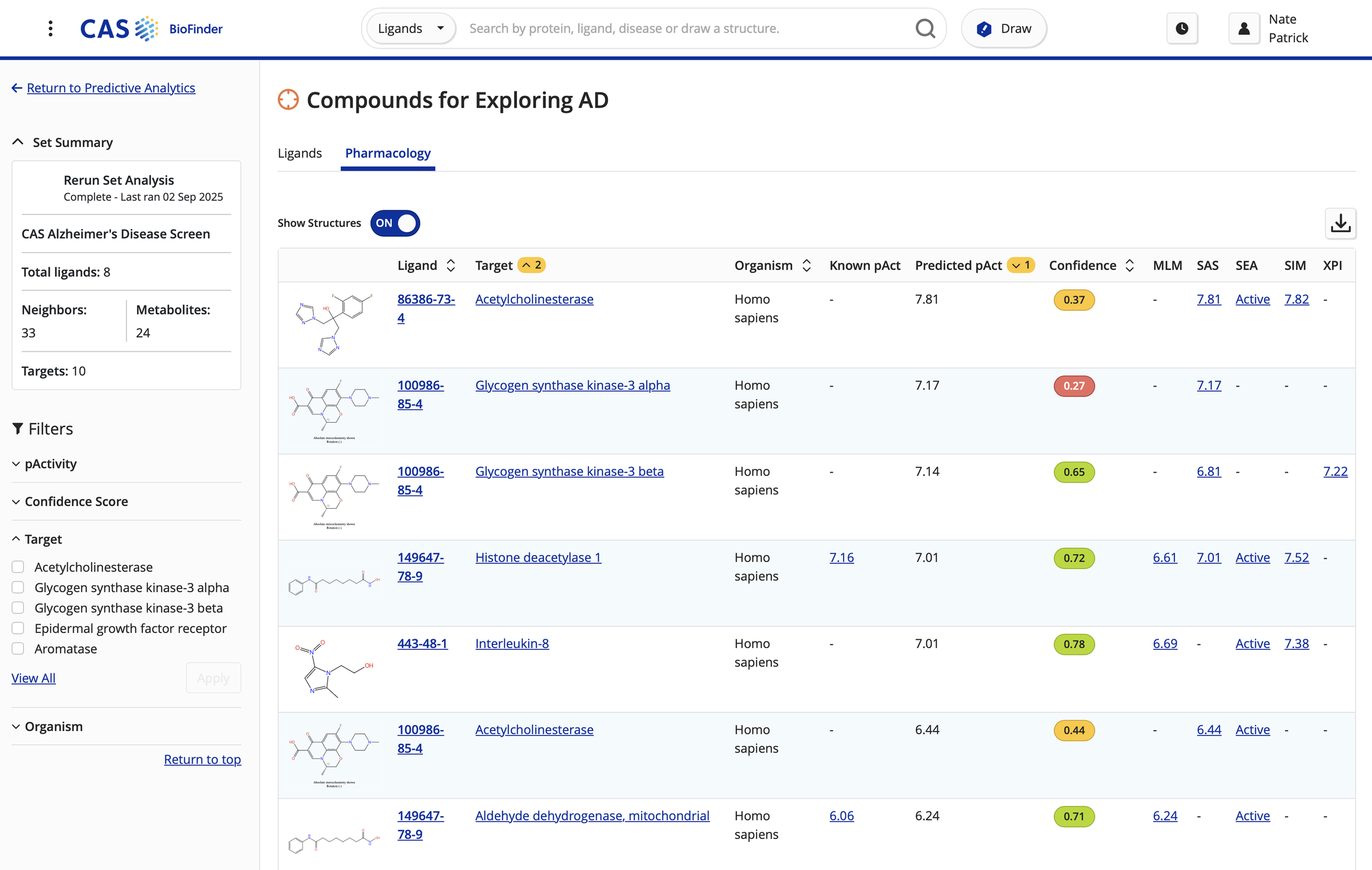
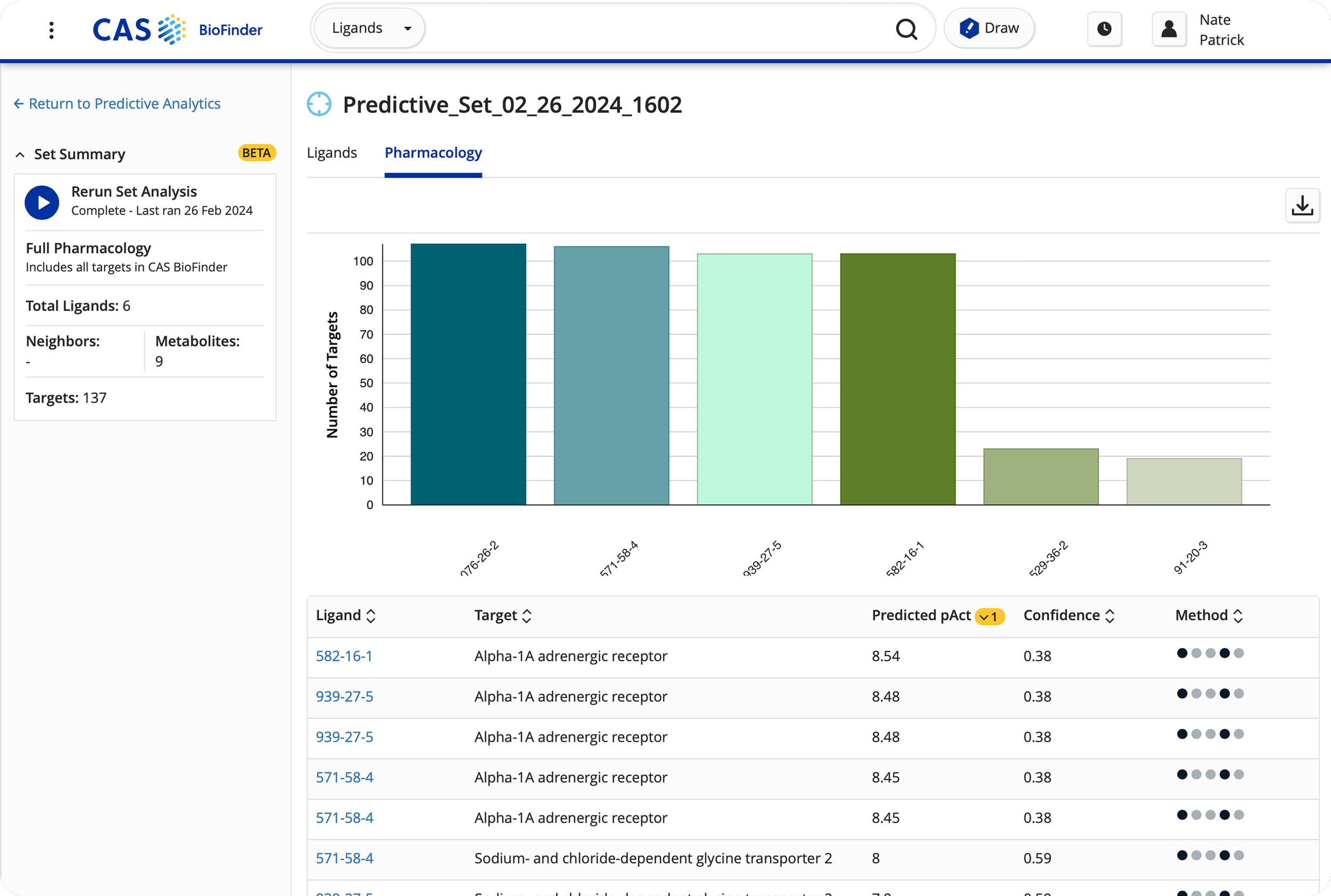
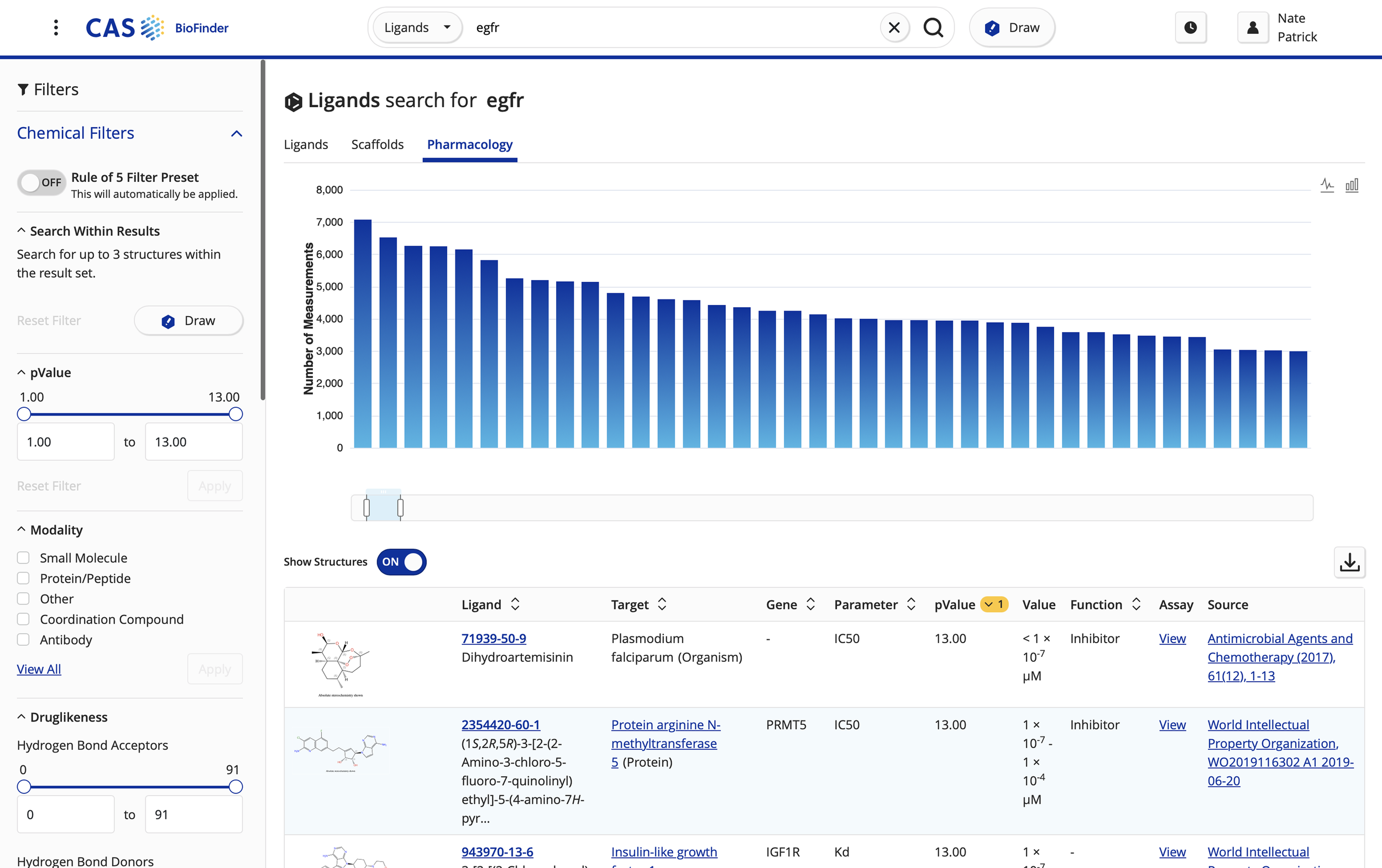
Designing activity analysis heat map to allow medicinal chemists to compare binding affinity of compared ligands against a target of interest.
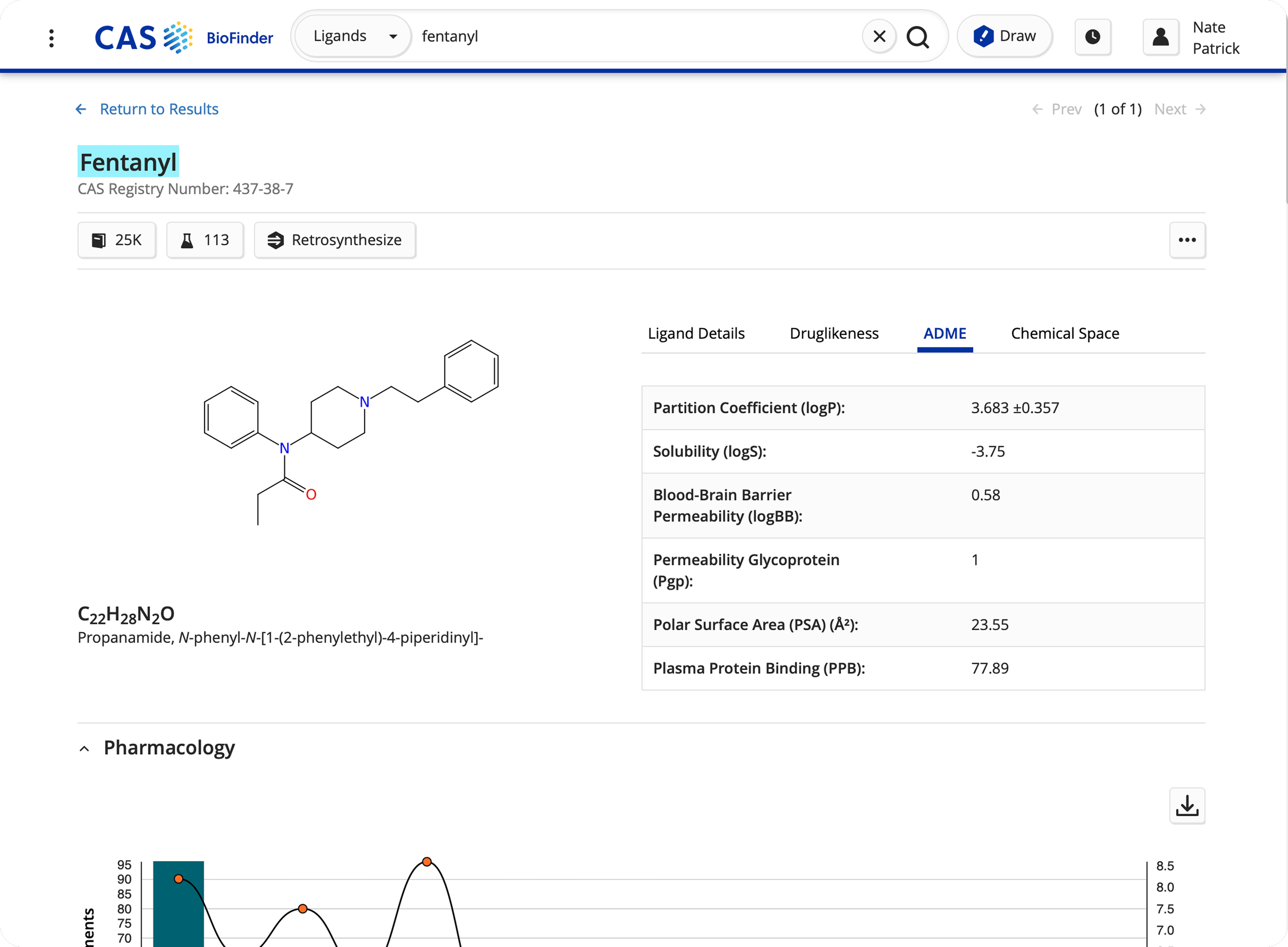
Revamped ligand/drug detail page for expansion of content types.
Refined predictive analytics feature to expand modeling capabilities, showing chemical structures next to protein/gene data to enhance data visualization.
Development of revamped sequence search capabilities to allow biologist personas to search proteins by sequence algorithms, directly linking them to available data on protein/gene detail pages.
Drug Intelligence to summarize FDA drug authorizations, dosages, and date of approvals.
CAS BioFinder homepage as of August 2025.
Version 2 Statistics
With the integration of first-time onboarding, and designing various automated flows which would give users a tour of the application, as well as the laundry-list of enhancements, traffic drastically increased in CAS BioFinder.
Over the course of the summer of 2025, sales would effectively close 12 major sales, including two academic consortia, opening up the use case for university usage. This resulted in 3.67 million in additional sales revenue across 14 countries.



![Chemical structure diagram of the compound 5-[3-(Phenylmethoxy)phenyl]-7-[trans-3-(1-pyrrolidiny methyl)cyclobutyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, with molecular details and drug information on the right side of the image.](https://images.squarespace-cdn.com/content/v1/5c266b8a3917eec9ff56251a/c982bd46-147a-44cc-aba3-faeb07c2dd75/image+48.png)